Rapid identification of potato cytoplasm types (Hosaka and Sanetomo 2012)
1. DNA extraction
Any DNA extraction method can be used. Here, FTA PlantSaver Card is used.
- Press plant tissue onto the card (FTA PlantSaver Card WB120065, Whatman). Allow to dry completely.
- Punch a 2.0 mm disk out of the FTA matrix impregnated with plant material using Harris Uni-core 2.0 mm Punch (WB100029, Whatman).
- Place the 2.0 mm disk in a 1.5 ml tube. Add 200 μl of FTA Purification Reagent, and leave for 5 min. Discard used reagent by a Pipetman.
- Add 200 μl of FTA Purification Reagent, and leave for 5 min. Discard used reagent by a Pipetman.
- Add 200 μl of isopropanol, and leave for 5 min. Discard used isopropanol by a Pipetman.
- Add 200 μl of isopropanol, and leave for 5 min. Discard used isopropanol by a Pipetman.
- Leave the lid open for several hours to dry the disk completely in a tube.
2. Multiplex PCR
- Drop a 2 mm dried disk into PCR tube, and then, dispense 5 μl each of the Reaction mix for FTA (or place 1 μl of DNA into the bottom of PCR tube, and then, dispense 4 μl each of Reaction mix).
- PCR [95°C 10 min, 35 cycles (94°C 30 sec, 60°C 30 sec, 72°C 1.5 min), 72°C 5 min, soak at 4°C]
| Component | Volume | Reaction mix (xn) | Reaction mix for FTA (xn) |
|---|---|---|---|
| DNA (5 ng/μl) | 1 μl | - | - |
| Dried FTA disk | - | - | - |
| 2x Ampdirect Plus | 2.5 μl (1x) | 2.5 x n μl | 2.5 x n μl |
| BIOTAQ HS DNA Polymerase (5 units/μl) | 0.025 μl (0.125 units) | 0.025 x n μl | 0.025 x n μl |
| 10x Primer mix | 0.5 μl | 0.5 x n μl | 0.5 x n μl |
| Sterile water | 0.975 μl | 0.975 x n μl | 1.975 x n μl |
| Total | 5 μl | 4 x n μl | 5 x n μl |
| Marker | Primer (5' - 3' sequence) | Conc. (μM) |
|---|---|---|
| T | GGAGGGGTTTTTCTTGGTTG AAGTTTACTCACGGCAATCG | 2 |
| S | GGTTCGAATCCTTCCGTC GATTCTTTCGCATCTCGATTC | 2 |
| SAC | TTGGAGTTGTTGCGAATGAG GTTCCCTAGCCACGATTCTG | 2 |
| D | CGGGAGGTGGTGTACTTTCT ACGGCTGACTGTGTGTTTGA | 3 |
| A | AACTTTTTGAACTCTATTCCTTAATTG ACGCTTCATTAGCCCATACC | 3 |
3. BamHI digestion
- After PCR, add 5 μl of the following Digestion mix.
Component Volume Digestion mix (xn) 10x NE Buffer 3 (New England Biolabs) 1 μl 1 x n μl 100x BSA (10 mg/ml, New England Biolabs) 0.1 μl 0.1 x n μl BamHI (20 units/μl, New England Biolabs) 0.3 μl (6 units) 0.3 x n μl Sterile water 3.6 μl 3.6 x n μl Total 5 μl 5 x n μl
- Mix the reaction by vortexing and spin down. Incubate at 37°C for over 3 hr in a PCR machine.
or,
- After PCR, add 5 μl of the following Digestion mix.
Component Volume Digestion mix (xn) 10x Fast Digest (Fermentas) 1 μl 1 x n μl FastDigest BamHI (10 units/μl, Fermentas) 0.5 μl (5 units) 0.5 x n μl Sterile water 3.5 μl 3.5 x n μl Total 5 μl 5 x n μl
- Mix the reaction by vortexing and spin down. Incubate at 37°C for over 5 min in a PCR machine.
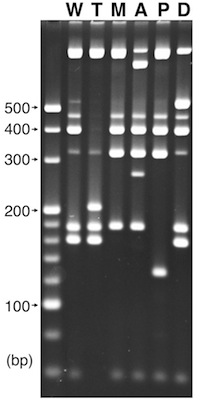
4. Agarose gel electrophoresis
- Mix with 3 μl of PCR products loading dye (sterile water: 10x Dye = 17:13) (10x dye = 0.25% bromophenol blue, 0.25% xylene cyanol, and 25% Ficoll Type 400).
- Electrophoresis in a 3% agarose gel in 1x TBE buffer (89 mM Tris-borate, 89 mM boric acid, and 2 mM EDTA).
Note
- 2x Ampdirect Plus can be replaced by standard PCR buffer and dNTPs.
- BIOTAQ HS DNA Polymerase (5 units/μl) can be replaced by other Taq DNA polymerase (hot-start type is preferable).
- For extension at 72°C in PCR, 1.5 min is critical.
- Banding patterns of different cytoplasm types are shown on right.
